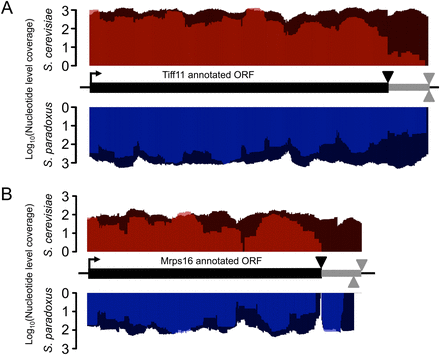
Evidence of stop-codon readthrough leading to C-terminal peptide extension. The translation initiation codons are indicated by the right-facing arrow, the annotated ORF by the thick black lines, and the canonical stop codon by the black triangles. The candidate C-terminal peptide extension is indicated by the gray line terminated by in-frame stop codons in the 3′ UTR (gray triangles above the line for S. cerevisiae, and below for S. paradoxus). Dark shades (red, S. cerevisiae; blue, S. paradoxus) indicate nucleotide-level coverage of mRNA fraction reads, and light shades indicate Ribo fraction reads. (A) Example of conserved C-terminal peptide extension of the translation initiation factor eIF1A (TIF11). The putative 21-amino-acid extension is conserved and well covered by reads in the Ribo fraction of both species. (B) Example of a S. paradoxus–specific C-terminal extension in MRPS16, a subunit of the mitochondrial ribosome. mRNA fraction reads indicate that the 3′ UTR is expressed in both species; however, translation is only detected in the 17-amino-acid extension of S. paradoxus, and not the potential 21-amino-acid extension of S. cerevisiae. Interestingly, coverage of the C-terminal extension in S. paradoxus is comparable to that of the CDS, suggesting that readthrough of this gene may be frequent.











