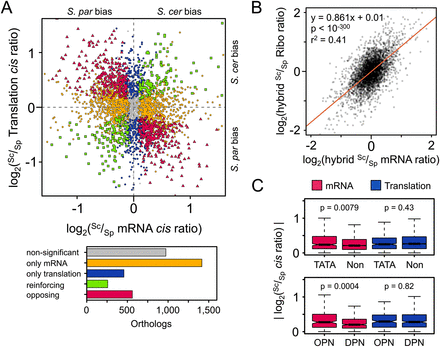
(A) The relationship between cis-regulatory divergence at the mRNA abundance and translational levels (all plotted Sc/Sp ratios are the mean of the two biological replicates). Divergence was detected only at the mRNA level for a large fraction of genes (orange circles), though greater than one-tenth of orthologs were significantly diverged only in translation (blue circles). Among orthologs diverged at both levels, we observed a significant excess opposing (red triangles) as compared with reinforcing changes (green squares). The number of orthologs in each class is indicated in the barplot. (S. cer) S. cerevisiae; (S. par) S. paradoxus. (B) Opposing divergence across regulatory levels. The red line indicates the best fit of a linear regression, with equation, p, and r2 values indicated above. The slope is significantly lower than one (95% confidence interval ±0.033), indicating that Sc/Sp mRNA ratio estimates tend to overestimate the degree of difference by ∼15% relative to that of the Ribo fraction. (C) Orthologs whose promoters contain either TATA boxes (TATA) or occupied proximal nucleosome regions (OPN) (Tirosh and Barkai 2008) show more divergence in cis only at the mRNA level when compared with non-TATA promoters (Non) or depleted proximal nucleosome regions (DPNs), respectively. Kruskal–Wallis test P-values are indicated above each fraction.











