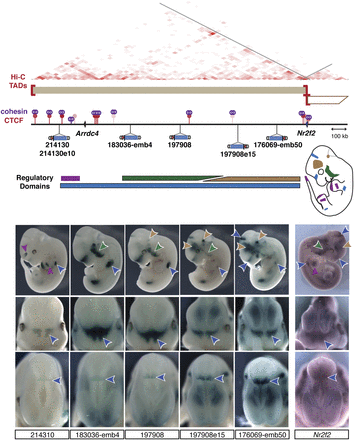
CTCF and cohesin sites are interspersed in regulatory domains. Schematic representation of the large topological/regulatory domain on chr7:75.5M–77.8M. The two genes (Arrdc4 and Nr2f2) are represented as arrows. The corresponding TAD is represented by a two-dimensional heat map (Dixon et al. 2012). Several constitutive CTCF sites (red lollipop, color intensity proportional to cell invariance), largely co-bound by cohesin (purple rings), are interspersed in this interval. Insertions spread across almost 2 Mb showed highly overlapping patterns in the proximal limb (blue arrow, top), face (blue arrow, middle), and at the midbrain/diencephalon boundary (blue arrow, bottom), forming a large regulatory domain. This large domain can be subdivided into smaller tissue-specific landscapes (green, purple, and brown) based on expression patterns displayed by only a subset of the insertions and quantitative differences in LacZ staining intensity. These different regulatory influences overlap with Nr2f2 expression, detected by whole-mount in situ hybridization. In situ hybridization with Arrdc4 probes did not reveal specific expression in E11.5 embryos. Embryos 183036-emb4 and 176069-emb50 were described previously (Ruf et al. 2011).











