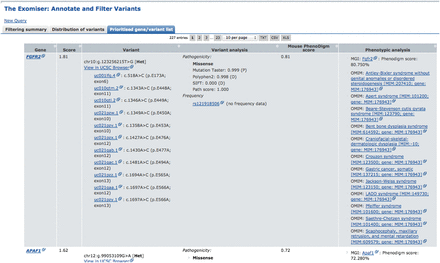
Exomiser querying of an exome containing a known chr10:g.123256215T>G heterozygous mutation associated with Pfeiffer syndrome (MIM:101600), an autosomal dominant Mendelian disease. The tab “Prioritised gene/variant list” shows the PHIVE prioritization of the 308 genes remaining after filtering of the original 8388 (details in Filtering summary table). The fully annotated variants associated with each gene, including pathogenicity and minor allele frequency, are shown along with the phenotypic relevance score from PhenoDigm and links out to any known phenotypic annotation from MGI/MGP or OMIM. The known variant is the top hit and annotated as a pathogenic, Glu to Ala missense coding change in FGFR2.











