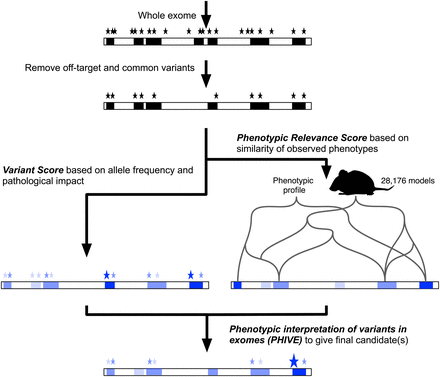
Exomiser filters a whole-exome data set by removing off-target, common, and synonymous variants from further consideration and evaluates the remaining variants based on the predicted pathogenicity and minor allele frequency (variant score). Optionally, an assumed mode of inheritance is used to further filter genes with variants present in a pattern compatible with the assumed mode of inheritance (e.g., homozygous or compound heterozygous for autosomal recessive). These genes are then assigned a phenotypic relevance score based on comparison with 28,176 mouse models with mutations in 9043 genes (7270 protein coding). The final ranking is calculated as the sum of the individual scores to yield the PHIVE score.











