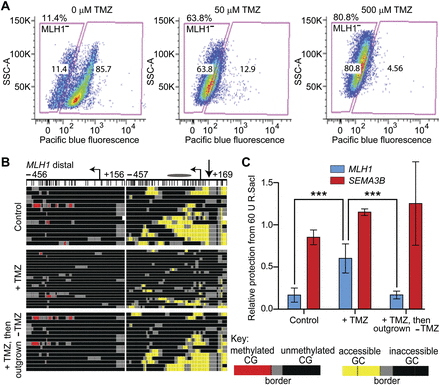
Cells with inaccessible chromatin at MLH1 are enriched upon treatment with TMZ. (A) Immunostaining with anti-MLH1 antibody and flow cytometry were conducted on GBM L0 cells after 72-h treatment with the indicated doses of TMZ. Chromatin accessibility at MLH1 was measured in control (top), TMZ-treated (+TMZ, middle), and TMZ-treated cells subsequently propagated in drug-free media (+TMZ, then outgrown –TMZ, bottom) by (B) MAPit-BGS (key at right) and by (C) protection from R.SacI activity. The location of the queried SacI site is indicated by the straight arrow next to the TSS in B (very top). Bars represent the mean protection from R.SacI activity for each locus ±SEM (control and +TMZ, n = 5; outgrown, n = 3), normalized to a control locus lacking a SacI site. A second control locus, SEMA3B, which contains a SacI site, but is inaccessible in GBM L0, was also assayed. (***) P < 0.001.











