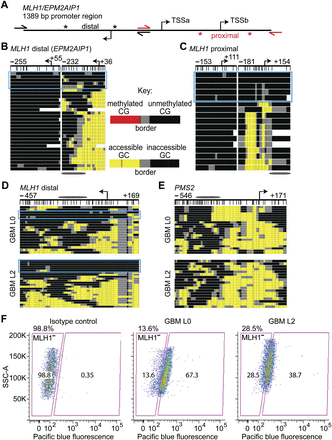
A subpopulation of molecules with relatively inaccessible chromatin at the MLH1 promoter is associated with MLH1-negative GBM cells. (A) Schematic of 1.4 kb of the MLH1 promoter. The three coregulated TSSs in this region are shown with bent arrows. Half-arrows indicate the primer binding sites for MLH1 distal (black) and proximal (red) MAPit-BGS amplicons. Asterisks indicate the boundaries of the MAPit-patch amplicons for the distal (black) and proximal (red) MLH1 promoter. MAPit-patch GC accessibility data is shown for the (B) distal and (C) proximal MLH1 promoter. Both amplicons show a subpopulation of relatively inaccessible molecules (circumscribed by cyan rectangles). MAPit-BGS GC accessibility at the (D) distal MLH1 and (E) PMS2 promoters in GBM L0 (top) and GBM L2 (bottom). Note the subpopulation of relatively inaccessible MLH1 molecules (enclosed by cyan rectangles). Schematics of the amplicon for the (D, very top) distal MLH1 promoter obtained using locus-specific primers (i.e., black half-arrows in A) and (E, top) PMS2 promoter are shown. An ellipse is shown scaled to 147 bp. (F) Immunostaining with an anti-MLH1 antibody and flow cytometry. (SSC-A) side scatter-A.











