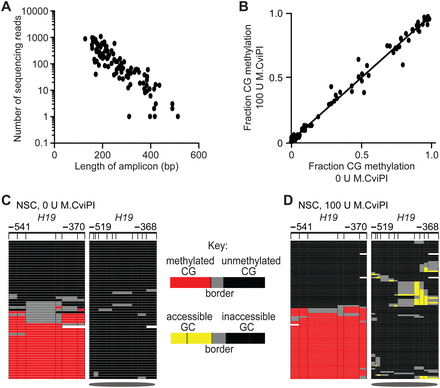
Probing chromatin with M.CviPI neither alters bisulfite patch PCR performance nor detection of CG methylation. (A) Number of sequencing reads decreases as a function of amplicon size. (B) Linear regression and Pearson's correlation plotted for CG methylation levels in NSC treated with 0-unit versus 100-unit M.CviPI. DNA methylation and chromatin accessibility at the imprinted H19 locus for NSC treated with (C) 0 units or (D) 100 units of M.CviPI. Symbols and the key for methylation status at right of C are as defined in Figure 1.











