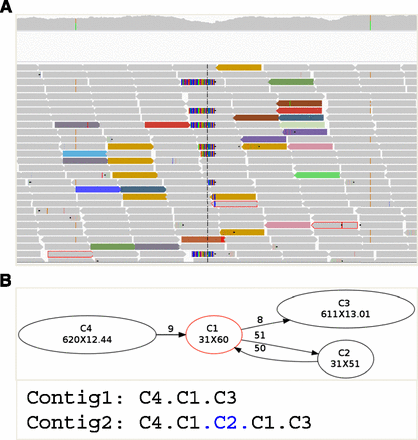
Schematic view of TIGRA. (A) Reads (arrow-shaped boxes) at a breakpoint (vertical dashed line in the center), including those normally mapped (gray), mate-unmapped (gray with red outline), soft-clipped (multicolored), and interchromosomally mapped (colored) are extracted from BAM files and sent to the assembly algorithm. (B) A de Bruijn graph is constructed using an iterative multiple-k-mer assembly algorithm. A contig (oval indexed node) with a specified length and average k-mer coverage (x) is connected to other contigs if it overlaps other contigs by k-1 bp (edge) in a particular orientation (arrow), and is of a particular coverage (weight). In this example, a mobile element insertion (of C2) with homology regions (C1) is successfully assembled. Two contig strings are decoded from the graph by TIGRA, representing two alternative alleles.











