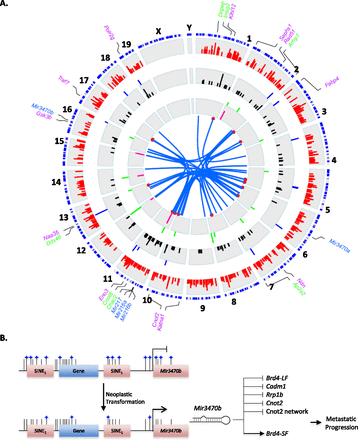
Integration of systems genetics analysis. (A) Circos integration of global transcript and miRNA profiling analyses. From periphery inward: clockwise proximal to distal chromosomal positioning (blue), (1) mRNAs showing statistically significant eQTL peaks (red peaks), (2) mRNAs with expression significantly associated with metastatic propensity (blue peaks), (3) hub transcripts encompassed within eQTL intervals (black peaks), (4) miRNAs encompassed within miR-eQTL intervals (green peaks), (5) miRNAs with metastasis-associated expression (pink peaks), (7) miRNAs predicted to target highly connected network hubs (blue line links miRNA and its predicted target mRNA, red point indicates mRNA position). Genes labeled in purple indicate module central nodes. Genes labeled in green indicate hub transcripts with predicted miR-3470b MREs that are down-regulated upon Mir3470b overexpression. miRNAs shown to have a causal role in metastasis are indicated in blue. Peak lengths are in −log10 P-value. Please refer to Supplemental Figure 44 for a detailed high-resolution version. (B) Proposed model for murine metastatic progression. Global genome hypomethylation associated with neoplastic transformation results in the DNA demethylation and transcriptional activation of short interspersed elements (SINEs) including Mir3470b. Mir3470b expression down-regulates metastasis suppressive genes and the metastatic progression inhibitory Cnot2 network while concurrently up-regulating the metastasis promoting Brd4-SF, driving metastatic progression.











