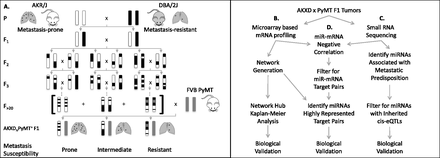
Genome-wide strategy to identify metastasis susceptibility co-expressed networks and their post-transcriptional regulators. (A) The pre-existing AKXD recombinant inbred (RI) panel was constructed by breeding metastasis-prone inbred mice from the AKR/J background to metastasis-resistant DBA/2J mice. F1 progeny from the AKR/J and DBA/2J cross were intercrossed; F2 progeny were bred to homozygosity, generating 24 isogenic AKXD sublines. Crossing each AKXD subline to the MMTV-PyMT model for highly metastatic breast cancer revealed phenotypic diversity in metastatic susceptibility within the AKXD RI panel. (B) RNA purified from AKXD subline primary tumors was subjected to global mRNA profiling by microarray followed by WGCNA and Kaplan-Meier Analysis to identify metastasis-associated co-expressed transcriptional modules. (C) AKXD subline tumor RNA was also subjected to small RNA sequencing followed by miRNA expression-phenotype correlations and eQTL analysis to identify metastasis susceptibility miRNAs. (D) Workflow illustrating analytic integration of global mRNA and miRNA profiling methods to uncover candidate post-transcriptional regulators of co-expressed networks.











