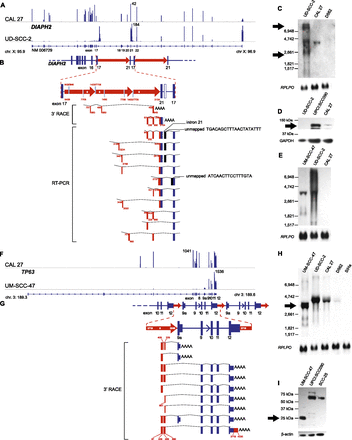
Focal HPV-associated genomic instability disrupts gene expression by diverse mechanisms. Disruption of DIAPH2 in UD-SCC-2 cells (A–E) and of TP63 in UM-SCC-47 cells (F–I) by HPV-associated genomic variation. (A) Mapped RNA-seq read histograms show 5′ expression of DIAPH2 transcript (RefSeq NM006729) in control CAL 27 (top) but not in UD-SCC-2 cells (bottom). Maximal read counts, chromosomal coordinates, and exon numbers are indicated. Truncated 3′ transcripts are overexpressed by fourfold in UD-SCC-2 cells. (B) Resolved linear structure of DIAPH2 locus after HPV integration, displaying (top and inset, middle; red arrows) direction of HPV integrants, not drawn to scale; (red and white numbers) HPV coordinates. (Bottom) Mapped viral–host fusion transcripts identified by 3′ RACE and RT-PCR. (C) Northern blot probed against DIAPH2 3′ cDNA demonstrates aberrant transcripts (arrows) in comparison to CAL 27 and UPCI:SCC090 controls. (D) Western blot showing that the protein translated from DIAPH2, i.e., diaphanous-related formin 2, is markedly reduced compared with control (top); GAPDH loading control (bottom). (E) Northern blot probed against HPV16 E7 demonstrating fusion transcripts in UD-SCC-2 and UM-SCC-47. (F) Histograms of mapped RNA-seq reads show expression of the 5′ end of TP63 in control CAL 27 but not in UM-SCC-47 cells. (G) Resolved linear structure of TP63 locus after HPV integration (top), not drawn to scale. Viral–host fusion transcripts detected by 3′ RACE (bottom) demonstrate HPV-associated gene disruption. (H) Northern blot probed against the 3′ end of TP63, showing truncated transcripts in UM-SCC-47 cells. (I) Western blot showing aberrant expression of 25-kDa p63 carboxy-terminal protein in UM-SCC-47 in comparison to 67-kDa conventional protein in UPCI:SCC090 and SCC-25 cells; (bottom) beta-actin loading control. See also Supplemental Figures 10 and 11.











