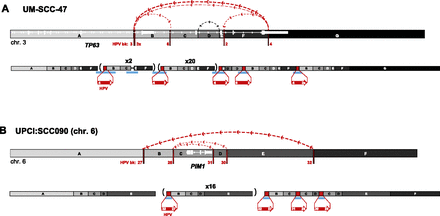
Connectivity maps of complex, HPV-associated genomic structural variation in HNSCC cell lines. Schematics of genomic target loci before (top) and after (bottom) HPV integration in HNSCC cell lines (A) UM-SCC-47 and (B) UPCI:SCC090, chromosome 6. Target gene schematic and connectivity map features are as described in the Figure 4 legend. HPV breakpoint numbers are listed in Supplemental Table 3. These schematics including the viral insertions are not drawn to scale. A short inversion in the viral sequence flanking breakpoint 3 is not shown. (Parentheses) Genomic segments with indicated fold amplification (x N) calculated from WGS data; (upside-down letters) inverted segments. See Supplemental Figure 9 for connectivity maps for both UD-SCC-2 and UPCI:SCC090, chromosome 9.











