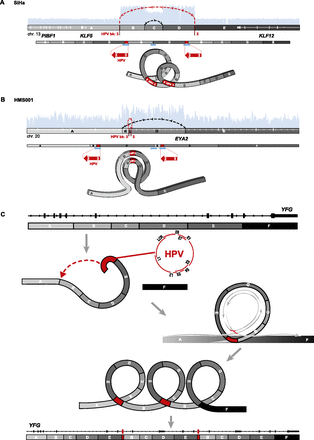
Focal amplifications and rearrangements explained by “looping” model. Schematics of HPV target sites before (top) and after (middle) viral integration in (A) SiHa and (B) HMS001 cells. We defined genomic segments (marked with capital letters and grayscale fills) based on HPV insertional (red lines) or host–host breakpoints (gray lines). (Top, white rectangles and horizontal lines) Gene exon structures; (arcing dotted lines, arrows: red) connections between HPV insertional breakpoints; (black) host–host breakpoints joining discontinuous reference genome sequences; (light blue histograms) depth of WGS coverage depicting CNVs (see Fig. 3). (Middle: direction of large red arrows) Relative orientation of sense strand of HPV reference genome, not drawn to scale; (white numbers and gaps) viral breakpoint ID numbers and rearrangements; (blue bars) confirmatory PCR amplicons and Sanger sequencing. (Bottom) Inferred, transient looping models to explain formation of CNVs and observed connections between breakpoints. (C) A generalized, stepwise looping model depicting HPV-associated structural variation at YFG (your favorite gene, top) as a target, inferred target nicking and integration of linear HPV genome (red), transient formation of circular DNA containing viral sequences, rolling circle amplification of this template (gray circular arrows), and formation of concatemers harboring identical viral–host and host–host breakpoints. The resulting observed focal amplifications and rearrangements, adjacent to HPV integrants, disrupt expression of YFG (bottom).











