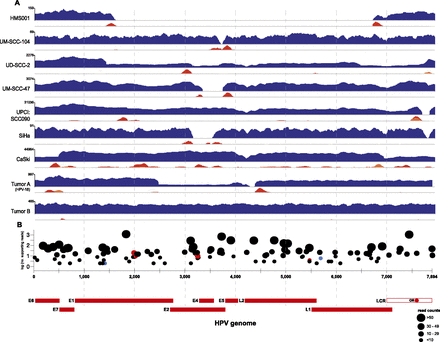
The HPV virome in human cancers. WGS reads were aligned to the HPV16 (x-axis coordinates, bottom) or HPV18 (Tumor A only) reference genomes. (A) In each row, histograms for various cancer samples indicate copy numbers of viral sequences (blue) and counts of discordant paired-end reads supporting insertional (red), intraviral (yellow), or both (orange) breakpoints. The scale (y-axis) of each plot was normalized to maximum read counts (upper left, each row). HPV breakpoints are defined further in Supplemental Figure 1 and Supplemental Table 3. (B) Verification of insertional breakpoints. Counts of discordant paired-end reads supporting each breakpoint (displayed both on the y-axis and by dot sizes), identified in any of the samples studied, are shown according to position in HPV16 genome (x-axis). (Black dots) Confirmed by PCR and Sanger sequencing; (red) failed PCR; (blue) not tested; (LCR) long control region; (OR) origin of replication. See also Supplemental Figure 2 and Supplemental Tables 3 and 4.











