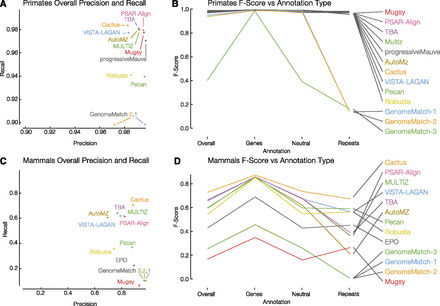
Simulated primate and mammal F-score results. Recall as a function of precision is shown for primates (A) and mammals (C). GenomeMatch-3 is omitted from plot A because both of its values are low (see its overall F-score in B). (B) The primate F-score results isolated to different annotation types: overall, genes, neutral and repetitive regions. (D) The mammal version of C. Legends for B and D are ordered as in the overall category and this order is maintained in the genes and neutral annotations.











