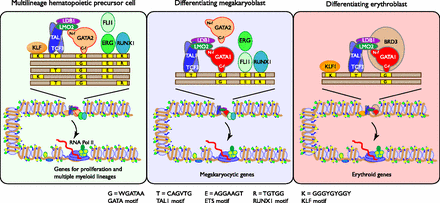
Model for differential occupancy by TAL1 in multilineage versus erythroid cells. The groups of proteins that co-occupy DNA in the two cell types are shown, along with cognate binding site motifs (yellow boxes, identity of each motif is listed at the bottom) along the DNA (long brown rectangle). The different arrangements of motifs signify the diversity of motifs seen in TAL1-occupied segments and emphasize the predominance of the GATA motif. The proteins TAL1:TCF3, LMO2, LDB1, and GATA1 form a multiprotein complex (Rodriguez et al. 2005); the other proteins shown are in proximity when bound to DNA. Additional proteins recognizing the same motif as other members of the TF family and bound at some sites (e.g., GATA2 and ERG in megakaryocytes) are also shown. Specific binding by TFs occurs within accessible DNA in chromatin that itself has histone modifications associated with gene activity (green and yellow circles representing methylation of lysines on H3 and acetylation of H4). The complex of proteins including TAL1 can exert positive effects on expression (curved arrow as shown) on recruitment and release of RNA polymerase for active transcription on induced genes. Other TAL1-containing complexes can exert negative effects (not shown). Cell-specific binding of TAL1 and associated proteins target different cohorts of genes.











