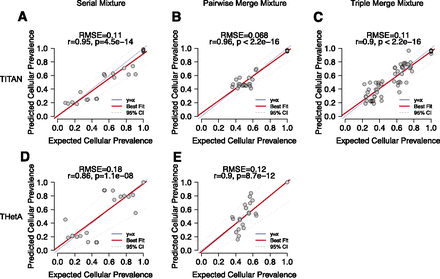
Performance of TITAN tumor cellular prevalence estimates for serial (30×) and pairwise (60×)/triplet (90×) merging simulations of intratumor samples from a HGS ovarian carcinoma. Pearson correlation coefficients (r) and root mean squared error (RMSE) were computed for TITAN (A–C) and THetA (Oesper et al. 2013) (D,E). Correlation and RMSE were computed by comparing the cellular prevalences of the predicted clusters with the prevalence of the expected clusters across the mixture samples. Each data point represents an expected clonal cluster with a unique tumor cellular prevalence. Ground truth and expected tumor cellular prevalence values were computed from the tumor contribution from each individual sample making up the simulated mixture (Supplemental Table 3B–D).











