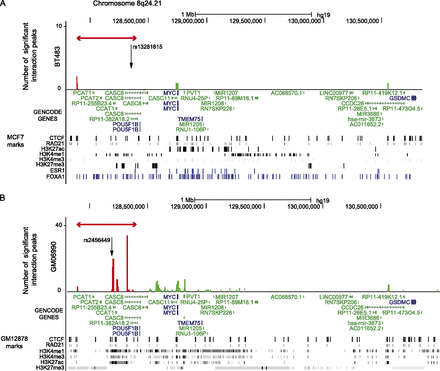
Statistically significant CHi-C interaction peaks at the 8q24.21 locus. (A) The number of statistically significant interaction peaks mapping to each HindIII fragment (y-axis) is plotted against the genomic location of the HindIII fragment (x-axis) for a 3.2-Mb region (127.8–131.0 Mb) of chromosome 8q24.21, including the 0.6-Mb genomic region (127,888,336–128,469,498 bp) that was targeted in the sequence capture step of our CHi-C protocol. All coordinates are based on hg19. The capture region is denoted by a double-headed red arrow; the “bait end” of the interaction peaks is indicated in red; the target end is indicated in green. The significant interaction peaks are aligned with (1) GENCODE genes (v19) with protein-coding transcripts colored blue and noncoding transcripts colored green; and (2) CTCF and RAD21 binding sites, active (H3K27ac, H3K4me1, and H3K4me3) and repressive (H3K27me3) histone modification marks (in black), and ESR1 and FOXA1 binding sites (in blue) generated in the breast cancer cell line MCF7 by the ENCODE Project, Frietze et al. (2012), and Hurtado et al. (2011). The location of the breast cancer risk SNP rs13281615 is also shown. (B) As above, but based on statistically significant interaction peaks in the lymphoblastoid cell line GM06990 and aligned with ENCODE data from the lymphoblastoid cell line GM12878.











