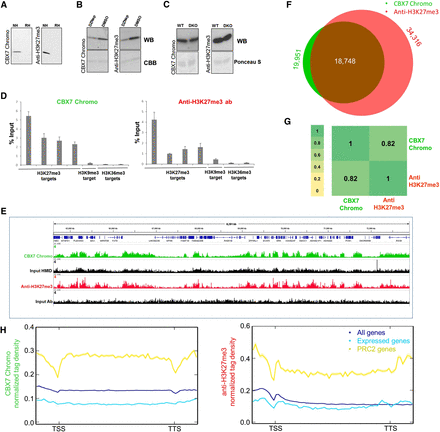
Comparative analysis of results obtained with the CBX7 Chromo domain and anti-H3K27me3 antibody No. 1 Lot 1 (Supplemental Fig. S1). (A) Western blot analysis of CBX7 Chromo and anti-H3K27me3 antibody binding to native (NH) and recombinant histones (RH). (B) Western blot analysis of CBX7 Chromo and anti-H3K27me3 antibody binding to nucleosomes isolated from EZH2 inhibitor (DZNep)- or DMSO-treated HepG2 cells. (C) Western blot analysis of CBX7 Chromo and anti-H3K27me3 antibody binding to histones isolated from wild-type and Suv39h DKO immortalized MEFs. (D) Comparison of the CIDOP-qPCR and ChIP-qPCR results of the CBX7 Chromo domain and anti-H3K27me3 antibody analyzed with amplicons associated with H3K27me3, H3K9me3, and H3K36me3. All experiments were carried out in triplicates of biological duplicates. Error bars represent the standard errors of the mean. (E) Representative genome browser snapshot of CIDOP-seq and ChIP-seq data obtained with the CBX7 Chromo domain and anti-H3K27me3 antibody (taken from ENCODE). For more examples, refer to Supplemental Figures S8C and S9. (F) Venn diagram of the overlap of peak regions between CBX7 Chromo domain and anti-H3K27me3 antibody. (G) Spearman correlation coefficient heatmap of CIDOP-seq and ChIP-seq tags in 10-kb bins. (H) Metagene profiles of CBX7 Chromo and anti-H3K27me3 antibody ranked by association with all genes, all expressed genes, and PRC2 (EZH2)-associated genes. The shaded lines represent the standard errors of the mean.











