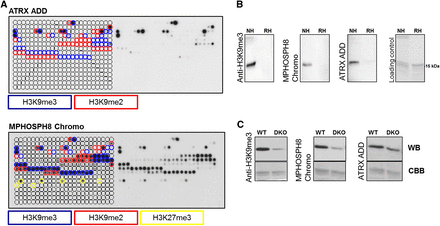
Peptide array and Western blot specificity analyses of H3K9me3-specific reagents. (A) CelluSpots peptide array analyses of ATRX ADD and MPHOSPH8 Chromo. Peptide spots are annotated on the left side of the glass slide. The color-coded boxes denote the presence of the designated modifications. ATRX ADD did not bind to H3K9me2/3 peptides also containing H3K4me2/3, H310Sph, or H3T11ph. Binding of MPHOSPH8 Chromo to peptides containing H3K9me2/3 or H3K27me3 was inhibited by H3S10ph, H3T11ph, or H3S28ph. For a more detailed annotation of the modifications at each spot, refer to Supplemental File S2. For comparison with anti-H3K9me3 antibodies, refer to Figure 1. The MPHOSPH8 data set was taken from Bock et al. (2011b) and reprocessed. (B) Western blot analysis using ATRX ADD, MPHOSPH8 Chromo, and anti-H3K9me3 antibody (Lot 2) with native (NH) and recombinant histones (RH). The approximate position of the H3 protein at 15 kDa is indicated. (C) Western blot (WB) analysis using ATRX ADD, MPHOSPH8 Chromo, and the same anti-H3K9me3 antibody with nucleosomes isolated from wild-type and Suv39h1 and Suv39h2 double knockout (DKO) cells. The Coomassie brilliant blue (CBB)-stained gel after transfer is shown as a loading control.











