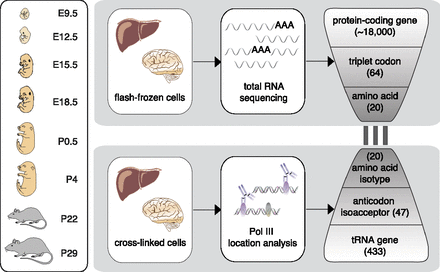
Transcriptome-wide analysis of protein-coding and tRNA genes during mouse organ development. Liver and brain tissues were isolated at eight mouse developmental stages. Tissue samples were flash-frozen for RNA-sequencing (RNA-seq) and cross-linked using formaldehyde to preserve protein–DNA interactions for ChIP-sequencing (ChIP-seq) of Pol III. Using the RNA-seq data, we calculated from all expressed protein-coding genes the frequencies of each triplet codon for all 64 possible codons and 20 amino acids. Similarly, Pol III binding to tRNA genes in the mouse genome was collapsed into 47 anticodon isoacceptor families and 20 amino acid isotypes (Methods). The bars linking RNA- and ChIP-seq data represent the three nucleotide interactions between codon and anticodon. Pol III occupancy was determined also in E9.5 (whole embryo) and E12.5 (head vs. remaining body).











