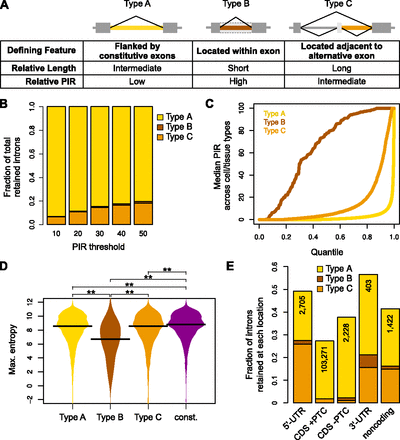
Distinct types of retained introns and associated properties. (A) Classification of retained introns and associated properties. (B) Fractions of total human retained introns belonging to each evolutionary type at different PIR thresholds. Represented are introns that could be assigned as Type A–C and that are retained at the indicated PIR thresholds in ≥10% of the samples. (C) Cumulative distribution of median PIR levels for each retained intron type in human cells and tissues. Only introns where PIR could be determined in ≥10% of the samples are represented. (D) Comparison of donor splice site strength (measured using maximum entropy; see Methods) of human retained introns and of constitutively spliced introns. Retained introns compared have PIR ≥ 10 in ≥10% of the samples where PIR could be determined; constitutively spliced introns have PIR < 2 in all samples where PIR could be determined. (Asterisks) P < 0.001 in two-sided Mann-Whitney U test. (E) Fraction of all human introns in each genic region that is retained with PIR ≥ 10 in ≥10% of the samples where PIR could be determined. (UTR) Untranslated region; (CDS) coding region of gene; (PTC) premature termination codon that can elicit nonsense-mediated mRNA decay. The total number of retained introns in each region is indicated.











