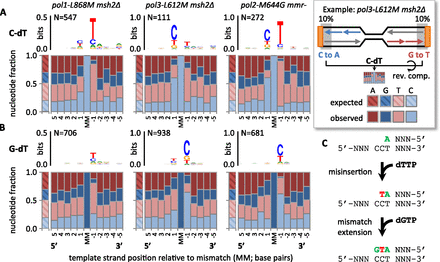
Sequence specificity of replication errors in the absence of MMR. (A) Nucleotide fractions and sequence logos (Schneider and Stephens 1990; Crooks et al. 2004) for five bases upstream of and downstream from mutations resulting from presumed C-dT mispairs, as calculated from sequences flanking mutations near replication origins (see example schematic). Expected fractions assume 38% G + C content. ([MM] Mismatch position; [N] mutation count pooled by strain.) (B) As per A, but for mutations resulting from presumed G-dT mispairs. (C) Example: An incoming mismatched nucleotide (red) stacks with adjacent pyrimidines (green) in the nascent strand, as indicated by logos in A.











