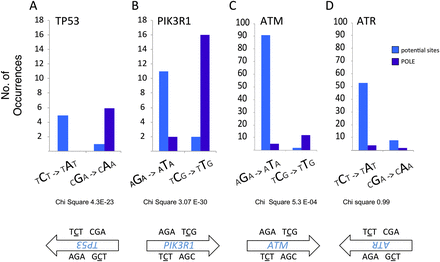
POLE-exo* mutations lead to enrichment of nonsense mutations at tumor suppressor genes in endometrial and colorectal tumors. Comparison of potential nonsense sites that occur in POLE-exo* context (TCT/AGA →TAT/ATA and TCG/CGA →TTG/CAA) to actual nonsense mutations found in POLE-exo* tumors. Selected tumor suppressor genes are shown. The number of potential nonsense mutation sites are shown in light blue; the nonsense mutation sites that actually occurred in POLE-exo* tumors are in dark blue. (A) TP53 (χ2 P = 4.3 × 10−23); (B) PIK3R1 (χ2 P = 3.07 × 10−30); (C) ATM (χ2 P = 5.3 × 10−4); (D) ATR; the POLE-exo* mutations are evenly distributed among the potential sites (χ2 P = 0.99). (See also Supplemental Tables S2, S3 for full amino acid change tables.)











