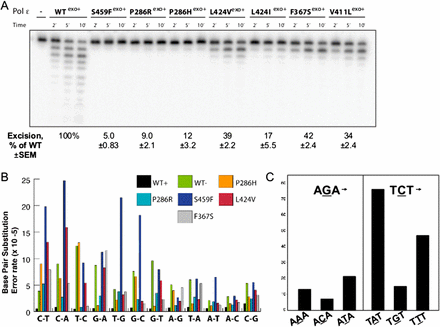
POLE-exo* cancer mutations severely and differentially inhibit 3′→5′ exonuclease activity in a strand-specific manner. (A) The relative 3′-5′ exonuclease activities of the POLE cancer mutant variants were compared to those of the wild-type POLE enzyme (WTexo+). Reactions were carried out at 37°C and initiated by the addition of the enzyme. Aliquots were removed at the indicated times and resolved by 12% denaturing PAGE (see Methods for reaction details). Exonuclease activity was quantified and expressed as a percentage of wild-type activity (±SEM). (B) POLE cancer variants selectively affect specific replication errors in vitro. Error rates for each of the 12 possible individual base pair substitutions were calculated using a lacZ forward mutation assay as described by Bebenek and Kunkel (1995) and Korona et al. (2011). (WT) A previously characterized exonuclease-inactivating mutant (D275A/E277A). (C) POLE-exo* P286R exonuclease mutants exhibit a preference for TCT→TAT mutations in a cell-free reaction. Purified POLE containing the P286R substitution, three equally spaced oligonucleotide primers, and an M13mp2 single-stranded DNA template were used to generate a sequence product that was directly sequenced using Illumina paired end protocol. In the resulting collection of sequenced POLE replication errors, the count of TCT→TAT mutations is higher than the count of the complementary AGA→ATA mutations. (See also Supplemental Fig. S3.)











