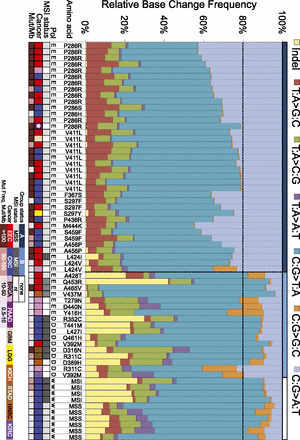
Mutation spectra of POLE and POLD1 exonuclease domain-mutated tumors. The color code for the type of mutation is shown in the key above the histogram. Tumors are grouped first by polymerase and then by affected residue with C→A frequencies decreasing from left to right. The darker blue designates Group A, medium royal blue Group B, and for comparison, white designates wild-type polymerase in MSI and MSS. Black horizontal dotted line demarks 20% C→A, the provisional threshold for classifying Group A. Black vertical line separates Group A from Group B, and wild-type polymerase categories. In addition, the horizontal bar at the top of the histogram shows the group designation. Cancer abbreviations are the following: (CRC) TCGA COAD/READ colorectal cancers; (EEC) endometrial cancer; (STAD) stomach adenocarcinoma; (GBM) glioblastoma; (LGG) low grade glioma; (BRCA) breast carcinomas; (PAAD) pancreatic adenocarcinoma; (HNSC) head and neck cancer; (KICH) kidney chromophobe. The MSI status, type of cancer, and mutation frequency range are color coded as shown in the key below the histogram. Other abbreviations are as follows: (MSS) microsatellite stable; (MSI) microsatellite instable; (nt) not tested. The white asterisk in the cancer track demarks a pancreatic tumor that had both a POLE-exoP286R and POLD1-exoR311H. See structural features of these mutations in Supplemental Figure S1.











