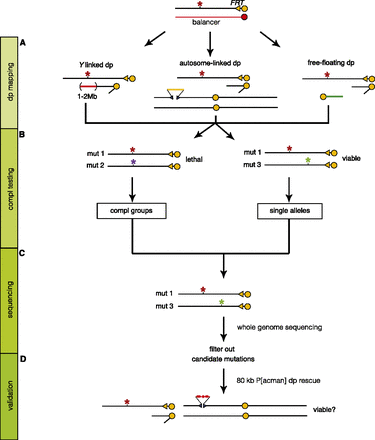
Mapping and sequencing strategy. General strategy to map lethal mutations on the X chromosome. (A) Duplication (Dp) mapping: For every mutant, lethality was mapped to an ∼1.4-Mb region by Dp mapping. (B) Complementation (Compl) testing: Mutations that map to the same duplication were intercrossed to identify Compl groups. (C) Sequencing: Whole-genome sequencing (WGS) was performed on a total of 394 transheterozygous mutations (mut 1/mut 3) whose lethalities map to a different duplication. The 394 mutations correspond to 258 single alleles and 68 complementation groups with two alleles. (D) Validation: We used 80-kb P[acman] duplications to rescue the lethality and confirm the mapping.











