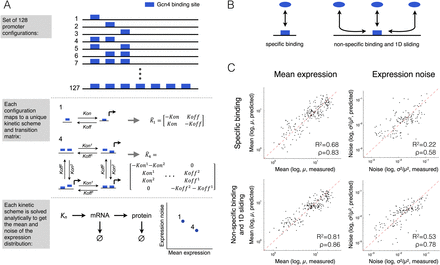
The effect of multiple transcription factor binding sites on noise is largely mediated by 1D sliding of the cognate transcription factor. For each promoter with a different number and configuration of seven possible TFBS (A, top), we constructed a kinetic model. Each of the 127 possible configurations was represented by a unique kinetic scheme and transition matrix (A, middle). The rate parameters of the reactions in the matrix were computed from the free parameters of the model using one of two alternative mappings that assume either that TFs search for their target through 3D diffusion (B, left) or that they do so by a combination of 3D and 1D diffusion (B, right). The transition matrix, together with reactions for transcription, translation, and mRNA and protein degradation, were simulated and solved analytically to obtain the steady-state mean protein abundance and noise (A, bottom). (C) The free parameters of the models were fitted in a leave-one-out cross-validation and the predictions (x-axis) of mean expression and CV2 were compared to the measured (y-axis) (see Supplemental Fig. S12 for a 10-fold cross-validation). The results show that the model that incorporates both 3D and 1D diffusion performs significantly better for both mean expression and noise than the model that assumes only 3D diffusion.











