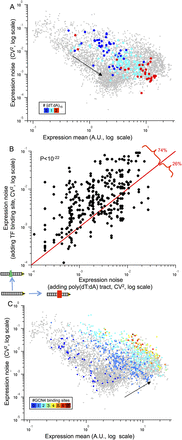
The effect of nucleosome-disfavoring sequences and number of binding sites on noise. (A) Mean expression and noise of 182 promoters with zero (blue points), one (light blue points), or two (red points) poly(dT:dA) of length 15 bp and a single Gcn4 site. Note that promoters with more poly(dT:dA) tend to have higher expression and lower noise. (B) Shown is a comparison of the effect on expression noise of increasing the mean expression either by adding a TF binding site or by adding a nucleosome-disfavoring sequence. Shown are 417 promoter pairs in which adding a TF binding site or a poly(dT:dA) tract resulted in a similar increase of the mean expression (all promoters were divided into 50 bins according to their log expression values, and promoters that share a bin are considered to have similar expression levels). In 309 (74%) of these 417 promoter pairs, adding a TF site resulted in noisier expression as compared to adding a nucleosome-disfavoring sequence (binomial test P < 10−22). (C) Mean expression and noise of 643 promoters with zero (dark blue points) to seven (dark red points) binding sites of Gcn4 and no other TF. The black arrows (A,C) point in the direction of expression and noise change. Note the general increase in both expression mean and noise that results in the addition of Gcn4 binding sites.











