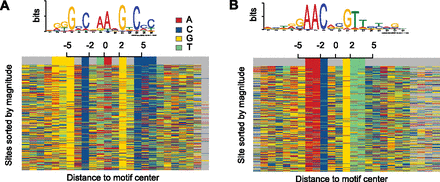
The binding motif can be observed in the sequence that surrounds the predicted binding site. This figure depicts the high resolution of our method. Each row indicates the sequence surrounding a predicted motif center. Different colors represent a different DNA letter. Results are shown for two distinct ChIP-seq experiments performed for the M. tuberculosis transcription factor DosR (A) and Kstr (B). A small shift at the motif center was allowed to improve visualization. Average shift was < 2 bp and is visualized by gray colors.











