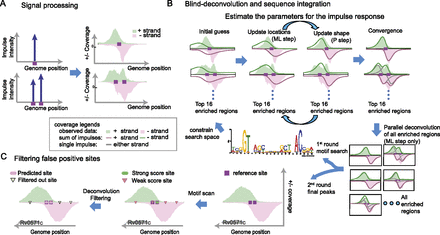
Illustration of the integrated model used to detect binding sites at high resolution. (A) The binding sites are a signal source. Each binding site (purple box) may emit an impulse response (blue upward arrow) that can be observed in the coverage of the ChIP-seq data (right). If two sites are close to each other, the observed data shows an overlap of the impulse responses from each site. (B) Illustration of the algorithm for binding site detection. The blind-deconvolution algorithm is broken into two parts to optimize the computational efficiency (see inset legend for detailed meaning of each line and color). First, both the ML and P steps are applied in a subset of enriched regions to estimate the parameters for the impulse response (top). Following, the ML step predicts the binding site locations for all regions in parallel (bottom right). From the output of the deconvolution process, we are able to predict a binding motif. This motif predicts potential binding sites that constrain the search space for a second round of the blind-deconvolution algorithm. This representation also illustrates the fit of a Gumbel distribution (green/red solid lines) in the ChIP-seq coverage (green/red shaded area). (C) Our method filters out false positives detected by the motif scan. Motif scan predicts binding sites that do not correspond to a true physiological binding. Our algorithm is inclusive with respect to low-affinity sites and uses the ChIP-seq coverage to filter out false positives.











