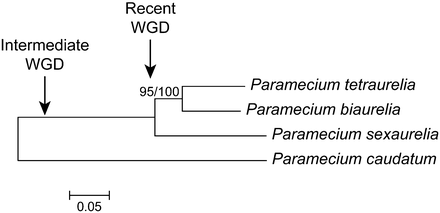
Maximum-likelihood phylogeny of P. biaurelia, P. tetraurelia, and P. sexaurelia, based on the protein sequence of genes predicted in this study (see Methods). P. caudatum was added as a preduplication outgroup (data for P. caudatum comes from McGrath et al. 2014). Approximate locations of recent and intermediate WGDs are indicated. Bootstrap value out of 100 replicates for the P. biaurelia–P. tetraurelia node is indicated. Scale bar represents 0.05 amino acid substitutions per site.











