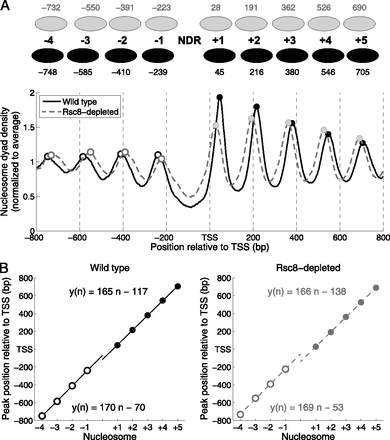
Nucleosome phasing relative to the transcription start site (TSS) shows global shifts of both upstream and downstream nucleosomal arrays toward the NDR in Rsc8-depleted cells, with no significant change in nucleosome spacing within each array. (A) Average dyad density near the TSS in wild-type (solid line) and Rsc8-depleted cells (dashed line). The average profiles were smoothed using a moving average filter with a span of 21 bp. The first four maxima upstream of and the first five maxima downstream from the TSS are marked by open and filled circles, respectively. The midpoints (dyads) of all nucleosome sequences between 140 and 160 bp were determined with respect to the TSS and summed for all genes. The data were normalized internally to the average value for each data set. (B) Linear functions fitted to the locations of the upstream and downstream maxima for wild-type (solid lines) and Rsc8-depleted cells (dashed lines) from A. Downstream array (filled circles); upstream array (open circles). The slope of each line gives the average distance between nucleosomes. Coefficients of determination, R2, were >0.999.











