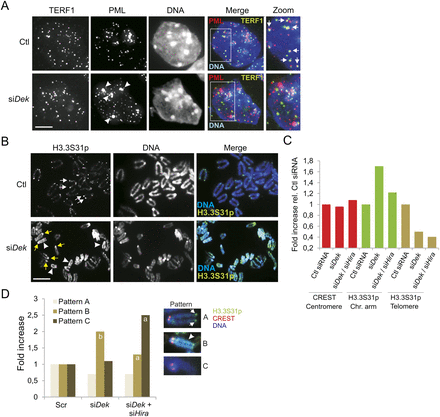
Loss of DEK in ESCs results in displacement of PML from telomeres, reduced H3.3 loading at telomeres, and HIRA-dependent H3.3 deposition on chromosome arms. (A) Immunolocalization of TERF1 and PML in control and DEK-depleted cells. Arrows in Ctl cells point to TERF1-PML colocalization; arrowheads in siDek cells show large PML NBs at sites of heterochromatin. (B) Immunofluorescence localization of H3.3S31p on mitotic chromosomes from control and DEK-depleted cells. Arrows in Ctl cells point to H3.3S31p at telomeres. White arrowheads in the siDek image show H3.3S31p on chromosome arms and yellow arrows show H3.3S31p at pericentromeres. Bars, 5 μm. (C) Fluorescence intensity level of CREST (as an invariant control marker; see panel D) and H3.3-EGFP on chromosome arms and centromeres in cells depleted of DEK or DEK + HIRA, relative to control cells (Ctl siRNA). (D) HIRA knockdown in DEK-depleted cells reduces H3.3S31p loading on chromosome arms. Graph shows the fold change in the proportion of metaphases with H3.3S31p patterns “A” (telomere staining; arrow), “B” (chromosome arm staining [arrowhead] and/or pericentromeric labeling), and “C” (no staining). Sixty mitotic spreads were examined. CREST was labeled as a centromere marker. P < 0.001 relative to Ctl and siDek; P < 0.001 relative to Ctl (Fisher’s exact test). Larger data sets are shown in Supplemental Fig. 6B.











