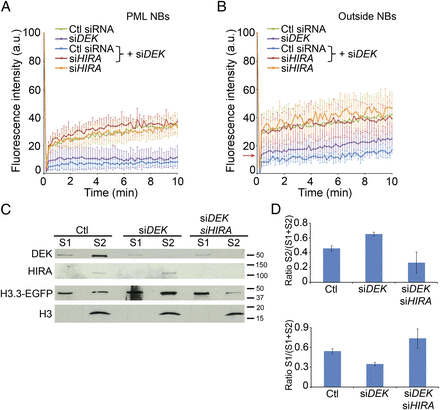
Depletion of DEK inhibits recruitment of H3.3-EGFP to PML NBs and promotes its loading on chromatin. (A) Loss of DEK by siRNA inhibits targeting of H3.3-EGFP to PML NBs. FRAP analysis of H3.3-EGFP at PML NBs 48 h after H3.3-EGFP transfection, in control and DEK-depleted cells (green and purple lines; mean ± SD of eight to 10 cells). H3.3-EGFP was also bleached in cells that were also depleted of HIRA as indicated (blue, red, and orange lines; mean ± SD of five to seven cells). Loss of HIRA restores the soluble pool of H3.3-EGFP sequestered after DEK depletion (cf. red and blue lines). (B) FRAP analysis of H3.3-EGFP outside NBs 48 h post-transfection in control and DEK-depleted cells. DEK depletion reduces the pool of soluble H3.3-EGFP (purple line; red arrow, recovery of soluble H3.3-EGFP 10 sec after bleaching) (nine to 14 cells). Cells were also depleted of HIRA as indicated (blue, red, and orange lines; five to six cells). (C) Western blot analysis of distribution of H3.3-EGFP in cells treated with control siRNA or siRNA to DEK or DEK + HIRA. Cells were fractionated into a 0.5% Triton X-100-soluble fraction (S1) and an MNase-soluble nucleosomal fraction (S2) (see also Supplemental Fig. 3C). (D) Densitometry analysis of H3.3-EGFP enrichment in S2 and S1 fractions (mean ± SD from duplicate experiments).











