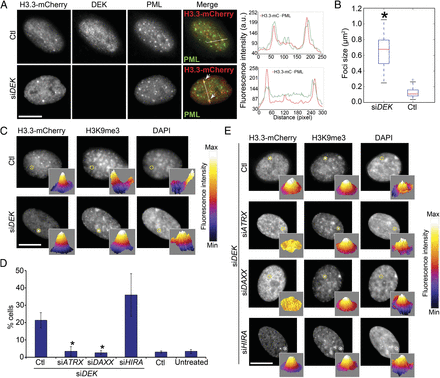
Depletion of DEK inhibits recruitment of H3.3-EGFP to PML NBs and promotes its loading on chromatin. (A) H3.3 forms large foci colocalizing with PML after DEK depletion. Localization of H3.3-mCherry, DEK, and PML in control (Ctl) and DEK-depleted (siDEK) cells, 48 h after H3.3-mC transfection. DEK was knocked down for 4 d by siRNA, at which time H3.3-mC was transfected concomitantly with a second round of DEK knockdown. (B) Median area of H3.3-mC foci in control and DEK-depleted cells. (*) P = 1.54 × 10−8 (Wilcoxon rank-sum test). Quantification of H3.3-mC foci as “large” arbitrarily required an area ≥0.3 µm2. (C) DEK depletion results in coenrichment of H3.3-mC foci in H3K9me3 and DAPI-dense DNA (100% of large H3.3-mC foci). Localization of H3.3-mC, H3K9me3, and DAPI intensity in control and DEK-depleted cells, with three-dimensional plots of H3.3-mC, H3K9me3, and DAPI intensity levels in control and DEK-depleted cells (circled area on the images). Scale: gray levels in arbitrary units; scale is identical within the marks analyzed. (D) Percentage of cells with H3.3-mC localized at heterochromatic foci marked by H3K9me3 in DEK-depleted cells also treated with siRNA to ATRX, DAXX, or HIRA. Data for control-transfected and untreated cells are also shown (right). (*) P < 10−4, Fisher’s exact test; mean ± SD of 200 cells per condition. (E) Localization of H3.3-mC, H3K9me3, and DAPI intensity in DEK-depleted cells also knocked down of ATRX, DAXX, or HIRA, with three-dimensional intensity plots as in C. Plots in C and E were generated using ImageJ. Note the lower H3.3 enrichment at these foci after knockdown of ATRX or DAXX, but not HIRA, despite the maintenance of the H3K9me3 and DAPI-dense marks. Bars, 10 µm.











