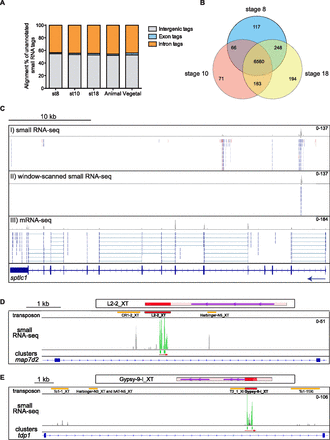
siteRNAs are conserved, cluster in introns, and align to TE remnants. (A) The percentage of unannotated small RNA tags that map to intergenic regions, exons, and introns (using RefSeq gene annotation) is shown for the stage 8, 10, 18, stage 10 animal, and stage 10 vegetal libraries. (B) Venn diagram showing the number and overlap of genes with small intronic noncoding RNAs in the stage 8, stage 10, and stage 18 libraries. (C) Small RNA alignments and mRNA-seq alignments (tracks I–III) to the sptlc1 gene at stage 10 shown in the Integrated Genome Viewer (IGV). Exons are indicated by dark blue boxes, introns by blue lines, and an arrow shows the direction of transcription. Small RNA tags are colored red for a sense alignment and blue for an antisense alignment to the genome, respectively. The gray histograms show peaks of read density with the scale indicated (e.g., 0–137 reads, track I). Track I shows small RNA-seq data before filtering by window-scanning analysis to reveal regions of high siteRNA density (track II). Track III shows mRNA reads, which align to exons. Reads that map over an exon–exon boundary are connected by a blue line. (D,E) Diagrams are shown for map7d2 and tdp1 to indicate the relationship between the siteRNA clusters and the TE remnants present in the introns. The representation of the genes is as in C. Small RNA reads are shown in the track labeled “small RNA-seq” in gray, or in green for those which passed the window scanning filter. The scale indicated is as in C. The green bars below indicate individual siteRNA clusters and the red arrow indicates the direction of transcription of the small RNA reads. In the track labeled “transposon,” the TE remnants are shown as colored boxes, with the names given below. The TE remnant from which the siteRNAs are derived is shown in red, and this entire TE is shown to scale above. The red box in the TE is the region of almost identical sequence to the intronic TE remnant. The purple line shows the protein-coding region and the arrow gives the direction of transcription of the entire transposon. Note that the small RNA reads are antisense relative to the direction of TE transcription.











