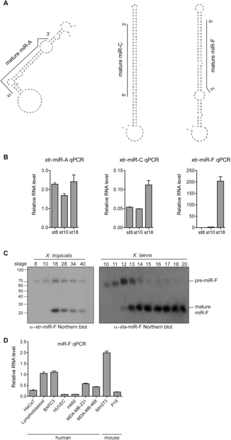
Identification of novel miRNAs. (A) Predicted pre-miRNA structures of X. tropicalis miR-A, C, and F. The black line indicates the mature miRNA sequence within the hairpin precursor. (B) qPCR detection of candidate novel miRNAs in X. tropicalis. Relative RNA levels to odc were calculated and error bars are standard deviations from triplicate repeats. (C) Northern blot for miR-F on RNA from X. tropicalis embryos of stages 8–40 (left panel) and X. laevis embryos of stages 10–20 (right panel). Both the mature miRNA (23 nt) and pre-miRNA (70 nt) are detected. Comparison to decade markers (Ambion) revealed the size of the small RNAs. (D) qPCR detection of miR-F in human and mouse cell lines. miR-F RNA levels relative to gapdh were calculated. Error bars are standard deviations of triplicate repeats.











