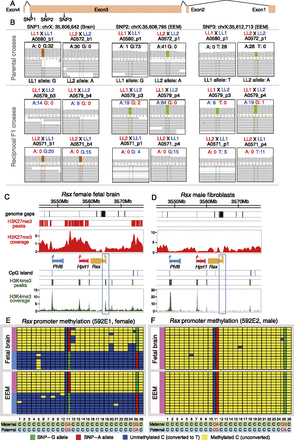
Allele-specific expression, allele-specific methylation, and histone modification profile for the noncoding RNA gene Rsx. (A) Exon model for the Rsx gene, with three SNPs between LL1 and LL2 opossum stocks indicated (blue arrowheads). (B) Allele-specific expression analysis of RNA-seq data for SNP1 in fetal brain and SNPs 2 and 3 in EEM in female offspring. Monoallelic paternal-allele expression was found at all three SNP positions in both tissues. (C,D) H3K4me3 and H3K27me3 histone modification profiles at the Rsx-containing region in female fetal brain (C) and control male fibroblast samples (D) from ChIP-seq experiments. In each panel, from top to bottom, are the genome gap locations (black bars), significant H3K27me3 peaks (red bars), H3K27me3 coverage (red scans), gene models, CpG island locations (blue bars), significant H3K4me3 peaks (green bars), and H3K4me3 coverage (green scans). (Blue) The escaper gene (Phf6) in this region; (red) the non-escaper gene (Hprt1); (orange) Rsx. (E,F) Bisulfite sequencing data for the Rsx promoter region in female (E) and male (F) fetal brain and EEM samples. The panel composition and colors are as described in Figure 6. Two SNPs found in this region (position 11 and 25) were genotyped in both parents by Sanger sequencing to infer the transmission direction and quantify methylation of the two alleles.











