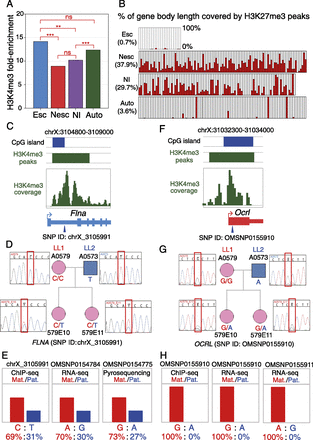
Histone modifications are correlated with maternal- versus paternal-allele expression of X-linked escaper and non-escaper genes. (A) H3K4me3 peak fold-enrichment for pXCI escaper genes (Esc), non-escaper genes (Nesc), X-linked genes with no informative SNPs (X-linked NI), and autosomal genes (Auto) in female fetal brain samples. Statistical significance was assessed by a Kolmogorov-Smirnov test: (ns) P-value > 0.05; (**) P-value < 0.01; (***) P-value < 0.001. (B) Barplot of percent H3K27me3 peak coverage in the gene region for four classes of X-linked genes in female fetal brain samples. Gene class abbreviations are as in panel A. Numbers in parentheses are average percent significant H3K27me3 peak coverage of all genes in the corresponding class. Among the X-linked NI genes, 11 had histone modification profiles similar to escaper genes and are designated as potential escapers of pXCI in opossum (Supplemental Table S7; Supplemental Text S3). (C–E) Allele-specific H3K4me3 modification for escaper gene Flna in female fetal brain ChIP-seq data from LL1 × LL2 cross. (C, top to bottom) CpG island location (blue bar), H3K4me3 peak/coverage profile (green bar and scan), 5′-end gene model. (Blue arrowhead) The position of a SNP (chrX_3105991) under the H3K4me3 peak with sufficient coverage to infer parent-of-origin allele-specific histone modification. (D) Sanger sequencing genotyping results for fetuses A0579E10 and A0579E11 (used for ChIP-seq experiments) and their parents confirmed that the SNP was informative in both F1 offspring. (E) Skewed H3K4me3 ChIP-seq and RNA-seq reads from the maternal allele and paternal allele at chrX_3105991 suggest activity of both alleles. The histone profile is consistent with the allele-specific expression profile at SNP OMSNP0154784 in the RNA-seq data and SNP OMSNP0154775 from the allele-specific pyrosequencing results. (F–H) Allele-specific H3K4me3 modification for non-escaper gene Ocrl in female fetal brain ChIP-seq data from LL1 × LL2 cross. (F) Features and color codes as for panel E. (Blue arrowhead) Position of SNP (OMSNP0155910) under the H3K4me3 peak with sufficient coverage to infer individual allele histone modifications. (G) Sanger sequencing genotyping results in the two embryos (A0579E10 and A0579E11) and their parents confirmed that the SNP was informative in both fetuses. (H) Unimodal H3K4me3 reads from the maternal allele at OMSNP0155910 suggesting that the on-mark is present only at the maternal allele. This is consistent with the maternal-specific expression at OMSNP0155910 and OMSNP0155911 in the RNA-seq data.











