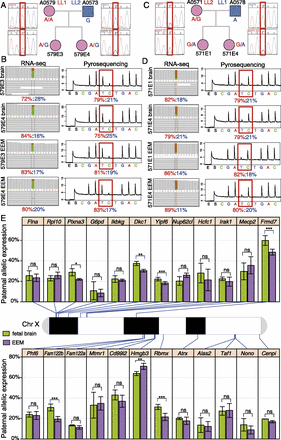
Maternal versus paternal allelic expression ratios for 24 pXCI escaper genes in opossum female fetal brain and EEM. (A–D) SNP genotyping and pyrosequencing verification for example escaper gene Yipf6 in opossum fetal brain and EEM. Sanger sequencing genotypes confirmed that exonic SNP OMSNP0155110 was informative in all four F1 embryos: (A) A0579E3 and A0579E4 in LL1 (dam) × LL2 (sire) cross; (B) A0571E1 and A0571E4 in the reciprocal F1 cross LL2 (dam) × LL1 (sire). (C,D) Biallelic expression verified by RNA-seq (left) and allele-specific pyrosequencing strategies (right). The results for all 24 escaper genes, a non-escaper gene, and an autosomal control gene can be found in Supplemental Figures S4–S28. (E) Names, physical locations, and allelic expression percentages for the 24 escaper genes identified from RNA-seq on the opossum X chromosome (centromere to the left). For each gene, the mean and standard deviation of paternal-allele expression, as a percentage of total expression, is plotted for fetal brain (green) and EEM (purple). Statistical significance, indicated by asterisks, was assessed by a Mann–Whitney U-test; (ns) P-value > 0.05; (*) P-value < 0.05; (**) P-value < 0.01; (***) P-value < 0.001.











