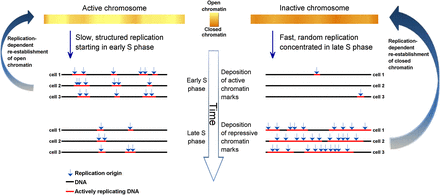
A model of replication structure regulation. Replication origins in regions that are transcriptionally active and have an open chromatin structure are activated at specified times (albeit with intercell variability in origin locations), giving rise to discrete domains that replicate in a synchronous manner. As S phase progresses, origin specification is gradually lost such that regions that replicate late use multiple origins are activated in a random manner, giving rise to an unstructured yet rapid replication pattern. In turn, dynamic changes in the activity of chromatin modifying enzymes over the course of S phase enable the preferential establishment of open or closed chromatin structures at particular genomic regions. The need to preserve chromatin structure during DNA replication may explain why cells sacrifice speed to achieve a specific temporal order of replication of transcriptionally active DNA.











