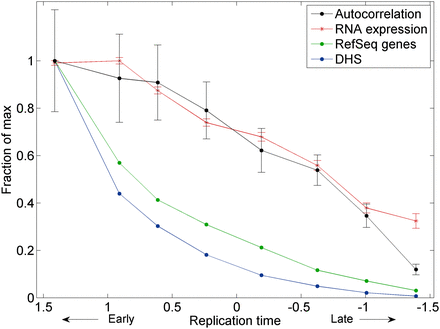
Random, unstructured replication of late-replicating, transcriptionally inactive genomic segments on the autosomes. Autosomal segments were divided into eight groups based on their replication timing. Shown for each group are the autocorrelation of replication timing at a ∼2-kb scale (averaged over all six individuals profiled; Methods), the number of RefSeq genes, average level of gene expression (measured by number of RNA-seq reads), and density of DNase I hypersensitive sites (DHS) representing the 5% most open chromatin regions in lymphoblastoid cell lines. Error bars, SE. All data are scaled relative to the maximal values for each data type.











