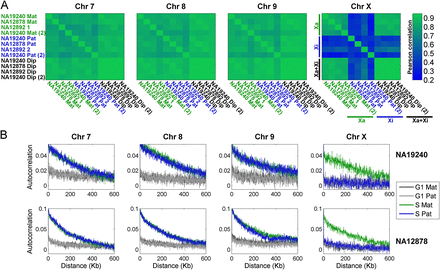
Structure and correlations of chromosomal replication profiles. (A) Correlation of replication timing for pairs of homologous chromosomes, within and between individuals and experimental replicates, shown as correlation matrices. For the autosomes, replication profiles were similar among individuals and between chromosome homologs in the same individual. Replication of the Xa was also consistent among individuals; however, replication of the Xi was different from the Xa, was uncorrelated among individuals, and was inconsistent between experimental replicates. (Mat) Maternal; (Pat) paternal; (1,2) homologous chromosome copies for which parent-of-origin is unknown (Supplemental Fig. S1); (Dip) diploid (aggregated across the two homologous chromosome copies); [(2)] experimental replicate. (B) Autocorrelations of DNA replication timing. The highly structured replication of the autosomes and the Xa is visible as long-range autocorrelation of DNA copy number in S phase genomes (but not in control, G1 phase genomes). In contrast, the inactive X chromosome shows no more autocorrelation in S phase than in G1 phase cells. Results for NA12892 were similar (data not shown). For both A and B, the 8-Mb left-distal part of the X chromosome was excluded from the analyses (see text), and results for other autosomes were similar (data not shown).











