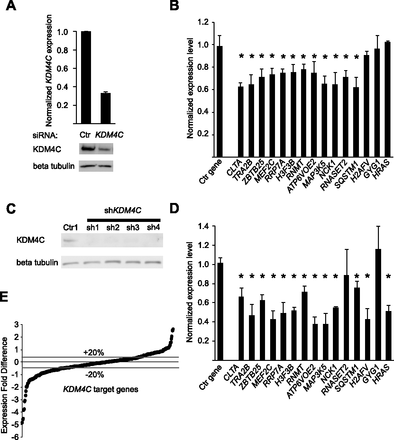
Molecular validation of KDM4C target genes in B cells and primary fibroblasts. (A) B cells from four individuals were treated with siRNA targeting KDM4C or a control siRNA. (Top) qRT-PCR data confirming KDM4C depletion at the transcript level; (bottom) a representative Western blot. (B) Gene expression changes in B cells from four individuals for 15 target genes, measured by qRT-PCR, following KDM4C knockdown. RPS6 is the control gene. Error bars, SEM ([*] P < 0.05, t-test). (C) Western blot of cells transfected with shRNA constructs targeting KDM4C or a control shRNA confirms knockdown of KDM4C in primary skin fibroblasts. (D) Gene expression changes of 15 target genes in primary fibroblasts by qRT-PCR following stable knockdown of KDM4C using the four shRNA constructs from C. RPS6 is the control gene. Ctr indicates control. Error bars, SEM ([*] P < 0.05, t-test). (E) Gene expression changes in 340 target genes measured by microarrays following KDM4C knockdown with sh1 (two biological replicates, in duplicate). Data from two additional shRNA constructs showed highly similar results (P < 0.001) (Supplemental Fig. S2B,C).











