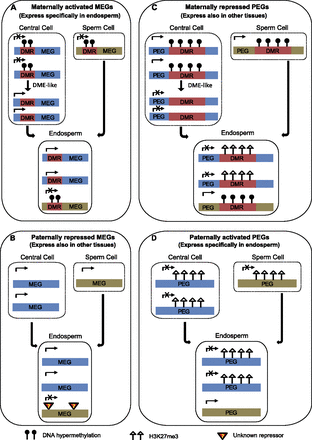
Models proposed for the regulation of imprinting in maize endosperm. (A) Model for maternally activated MEGs. MEGs expressing specifically in endosperm are probably maternally activated. Most of these MEGs are associated with pDMRs located around the upstream and 5′ portion of gene body regions. In central cells, the methylation of pDMR regions can be removed by DME-like demethylase, but not in sperm cells. After fertilization, pDMRs that were identified in the endosperm exhibit maternal DNA demethylation and paternal DNA hypermethylation. Maternal DNA demethylation of these MEGs results in their maternally preferred expression. (B) Model for paternally repressed MEGs. MEGs expressing in endosperm but also in other tissues are probably paternally repressed. The nonexpression of the paternal alleles of these MEGs may have resulted from paternally specific unknown repressor(s). (C) Model for maternally repressed PEGs. PEGs expressing in endosperm but also in other tissues are probably maternally repressed. Paternal preferred expressions of these PEGs require both DNA methylation and H3K27me3. In central cells, DNA methylation in pDMRs is removed, but not in sperm cells. In the endosperm, H3K27me3 only targeted to the maternal alleles with DNA demethylation of the pDMR regions (demonstrated by maternal-specific H3K27me3). As the paternal alleles remain hypermethylated, they cannot be targeted by H3K27me3. Repression of the maternal alleles by H3K27me3 led to the paternal preferred expression of these PEGs. (D) Model for paternally activated PEGs. PEGs expressing specifically in endosperm are probably paternally activated (de-repressed) PEGs. In other tissues, these PEGs possibly possess biallelic H3K27me3 peaks. As only maternal-specific H3K27me3 peaks were identified in endosperm, it is likely that for these PEGs, their paternal alleles have lost their H3K27me3, which resulted in paternally specific expression. The dashed boxes around the central cell and sperm cell indicate that the gene expression and chromatin modification in these tissues have not been supported by experimental data.











