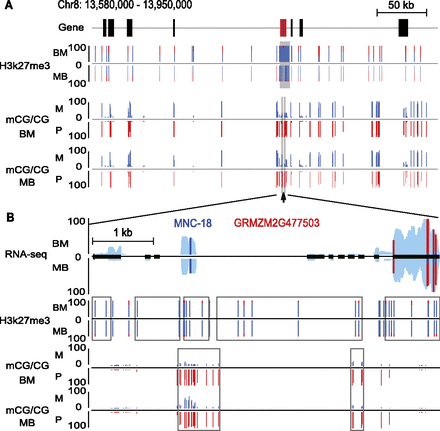
Allelic views of H3K27me3 and DNA methylation in a genomic region. (A) Allelic levels of H3K27me3 and CG DNA methylation are shown for an ∼400-kb region in both BM and MB endosperm. The region contained nine genes with unknown or nonimprinting status (black block), one PEG (red block), and one MNC (blue block). (B) A zoomed-in allelic view of RNA-seq, H3K27me3, and CG DNA methylation for a confirmed PEG that overlapped with a MNC. The overall expression level of transcribed regions is shown in light blue for both BM and MB. The relative expression levels, the percentage of allelic reads of H3K27me3 ChIP-seq data, and the DNA methylation level for specific SNP sites are shown for both maternal and paternal alleles, with red lines for the paternal allele (P) and blue lines for the maternal allele (M). Black rectangle, exon; black line, intron. The gray rectangles highlight the maternal H3K27me3 peaks and pDMRs identified in this region. The lines (or dots) correspond to all analyzable sites.











