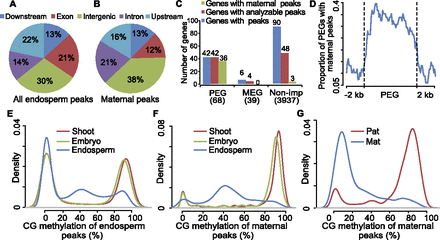
Analysis of maize H3K27me3 peaks. (A,B) Genomic distribution of 12,125 H3K27me3 peaks (A) and 1131 maternal peaks (B) identified from hybrid endosperm of reciprocal crosses. (C) Overlap between maternal peaks and MEGs/PEGs/nonimprinted genes (Non-imp). “Genes with peaks” indicates the genes that are overlapped with H3K27me3 peaks identified in the endosperm of both BM and MB. “Genes with analyzable peaks” refers to genes that had SNPs within the H3K27me3 enriched region. “Genes with maternal peaks” refers to genes that contained maternal-specific H3K27me3 peaks identified in this study. The number in parentheses indicates the number of genes that were analyzed. (D) The distribution of maternal-specific H3K27me3 peaks located in PEGs and their 2-kb up- and downstream regions. (E,F) The distributions of DNA methylation levels for maize endosperm H3K27me3 peaks (E) and maternal-specific peaks (F) in shoot, embryo, and endosperm of B73. (G) The distribution of parental DNA methylation of maternal peaks.











