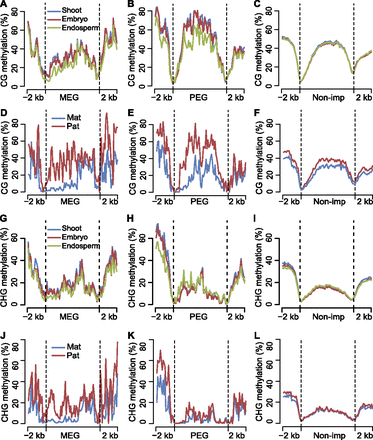
Differential DNA methylation among tissues and between two parental genomes for maternally and paternally expressed imprinted genes. (A–C) Average DNA methylation levels of MEGs, PEGs, and nonimprinted genes (Non-imp) for shoot, embryo, and endosperm in CG context throughout the gene body and its 2-kb up- and downstream regions. (D–F) Comparison of average DNA methylation levels between two parental genomes of MEGs, PEGs, and nonimprinted genes (Non-imp) in CG context throughout the gene body and its 2-kb up- and downstream regions. (G–I) Average DNA methylation levels of MEGs, PEGs, and nonimprinted genes (Non-imp) for shoot, embryo, and endosperm in CHG context throughout the gene body and its 2-kb up- and downstream regions. (J–L) Comparison of average DNA methylation levels between two parental genomes of MEGs, PEGs, and nonimprinted genes (Non-imp) in CHG context throughout the gene body and its 2-kb up- and downstream regions. (A–L) Gene body regions were separated into 60 bins, and extended 2-kb up- and downstream regions were separated into 20 bins. The average methylation levels were calculated with the same method as in Supplemental Figure S7.











